I have done two research internships applying network analysis to biological problems using high performance computing. Some of the details are given below. I hope to work on more such projects in the future.
My favorite things about applied work are getting to think about practical algorithmic efficiency, high performance computing and learning about new fields from experts. In the past I worked with people from bioinformatics, ecology, systems analysis and physics backgrounds.
The origins of life?!?!
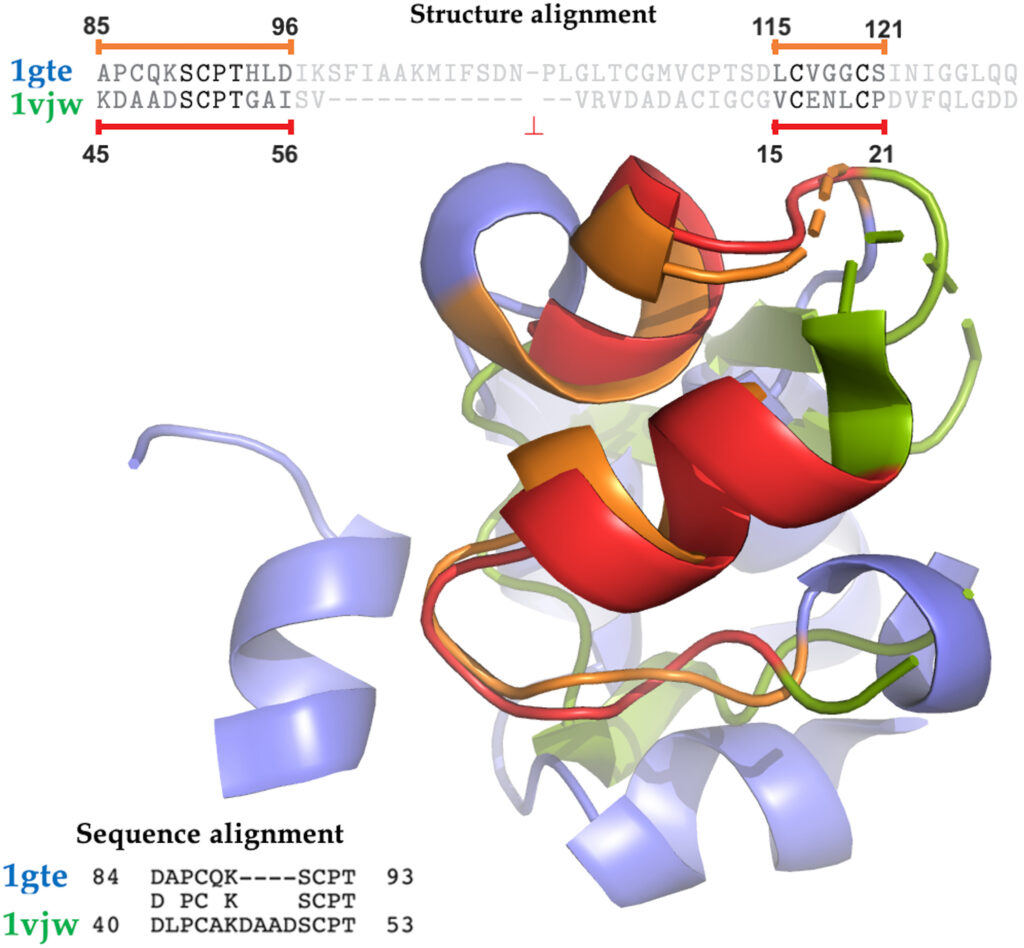
A project I worked on as intern in Professor Yana Bromberg’s lab has recently been published as a journal paper:
Quantifying Structural Relationships of Metal Binding Sites Suggests Origins of Biological Electron Transfer (Yana Bromberg, Ariel Aptekmann, Yannick Mahlich, Linda Cook, Stefan Senn, Maximilian Miller, Vikas Nanda, Diego Ferreiro, and Paul Falkowski). in Science Advances (an open access publication by the same publisher at Science)
Hand-wavy summary:
Energy is an organism’s most basic need. Organisms get energy by carrying out chemical reactions called “electron transfer” reactions. For example photosynthesis is an electron transfer reaction. Since since energy is so critical to life, our group wanted to better understand how organisms capacity for electron transfer reactions evolved.
- Here is non-technical news article discussing the paper.
- and an article in La Opinión!
- Here are some more detailed notes I wrote about the project and my work.
My contribution:
I was an intern at Yana Bromberg’s lab in bioinformatics while I was an undergrad at Rutgers. My primary job was to construct and analyze a protein structural similarity network to identify possible evolutionary relationships. The nodes in this network represented the structure of metal binding sites of proteins and the edges represented their structural similarity. Our dataset was taken from the Protein Data Bank. Since we had a huge number of nodes and we needed to know the structural similarity between each pair of nodes algorithm efficiency was critical! I used python, unix, and of course distributed computing to accomplish this task.

Aquatic Food Webs
In the summer of 2017, I was an intern at the Institute for Applied Systems Analysis in Laxenburg, Austria. I worked on the network analysis of aquatic food webs. My supervisors were Elena Rovenskaya from the Applied Systems Analysis group and Ulf Dieckmann from the Evolution and Ecology Program. My main contribution to the project was creating and running analysis code via high performance computing.
At IIASA, I was able use my expertise in structural graph theory and to study food web structures. In particular, I looked at the relationship between the distribution of motifs, the food web analog of an induced subgraph, and a food web’s vulnerability to ecosystem collapse following species extinction.
This project was funded by Princeton’s Mathematical Methods for Water Problems Project (NSF 1514606).
More about the the project:
While at IISAA, I helped analyze a database of quantified food webs, that is, networks of trophic ecological interactions, in which nodes are described by the material or energy content of populations of species or functional groups and directed edges are described by the strengths of the energy flows between those nodes resulting from feeding relationships. My work focused on the investigation of how different local interaction network structures called “motifs” are distributed in food webs. A major contribution was my development of analytic software tools to facilitate the analysis.
We had a database of over 200 food webs to analyze, each involving a large amount of relations. My biggest initiative was to run all scripts in parallel on Princeton’s super computer which was critical given the huge dataset. Prior to my arrival at the lab all computation had been done locally on a single computer. Switching to using distributed computing increased the scope of calculations we could run on our large dataset and allowed us to get results more quickly.





IIASA headquarters are pictured above.